Introduction
Statistical analysis of somatic hypermutation (SHM) patterns in B cell immunoglobulin (Ig) sequences often require background models of SHM. This website provides: (1) a targeting model that defines where mutations occur (by specifying the relative rates at which DNA motifs in the Ig sequence are mutated), and (2) a nucleotide substitution model that defines the resulting mutation (by specifying the probability of each base mutating to each of the other three possibilities as a function of the surrounding bases). These models are based on the analysis of many high-throughput Ig sequencing datasets using the methods developed in:
“A model of somatic hypermutation targeting and substitution based on synonymous mutations from high-throughput Immunoglobulin sequencing data” Gur Yaari, Jason Vander Heiden,, Mohamed Uduman, Daniel Gadala-Maria, Namita Gupta, Joel N.H. Stern, Kevin C. O'Connor, David A. Hafler, Uri Laserson, Francois Vigneault and Steven H. Kleinstein. 2013. (Frontiers in Immunology, Submitted)
The current “S5F” models (v07312013.1) are based on 806,860 synonymous mutations in 5-mer motifs from 1,145,182 Functional sequences from the following data sets:
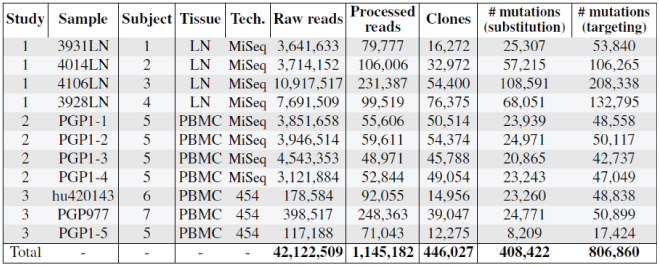
The models available through this website will be updated to include new data sets as they become available.
